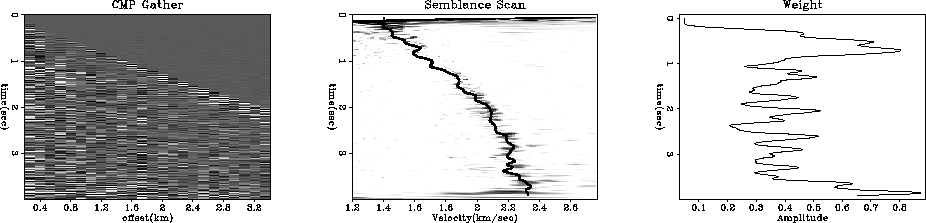
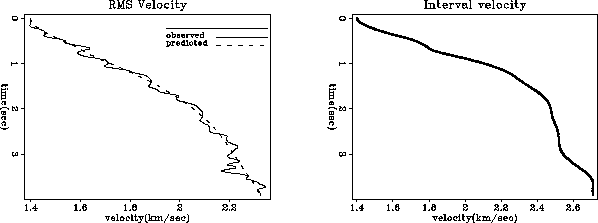
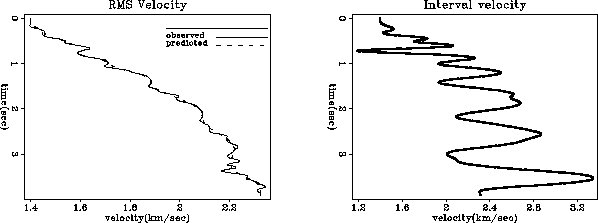
module vrms2int_mod { # Transform from RMS to interval velocity
use causint
use cgstep_mod
use solver_mod
contains
subroutine vrms2int( niter, eps, weight, vrms, vint) {
integer, intent( in) :: niter # iterations
real, intent( in) :: eps # scaling parameter
real, dimension(:), intent( in out) :: vrms # RMS velocity
real, dimension(:), intent( out) :: vint # interval velocity
real, dimension(:), pointer :: weight # data weighting
integer :: st,it,nt
logical, dimension( size( vint)) :: mask
real, dimension( size( vrms)) :: dat
nt = size( vrms)
do it= 1, nt {
dat( it) = vrms( it) * vrms( it) * it
weight( it) = weight( it)*(1./it) # decrease weight with time
}
mask = .false.; mask( 1) = .true. # constrain first point
vint = 0. ; vint( 1) = dat( 1)
call solver_prec( causint_lop, cgstep, x= vint, dat= dat, niter= niter,
known= mask, x0= vint, wt= weight,
prec=causint_lop, eps= eps, nprec= nt)
call cgstep_close()
st = causint_lop( .false., .false., vint, dat)
do it= 1, nt
vrms( it) = sqrt( dat( it)/it)
vint = sqrt( vint)
}
}