 |
 |
 |
 | Krylov space solver in Fortran 2009: Beta version |  |
![[pdf]](icons/pdf.png) |
Next: Conclusions
Up: Krylov space solver in
Previous: Solvers
Converting RMS velocities to interval velocities is one of the most basic problems
in reflection seismology. The Dix equation is one of the most common approaches
but often leads to unrealistic velocities when dealing with the noise in the RMS
velocity measurement. Clapp et al. (1998) points out that there is a linear
relationship between
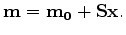 and
and
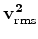 using either a modified
version of causal integration or using causal integration
using either a modified
version of causal integration or using causal integration 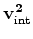 directly and first
scaling
directly and first
scaling
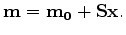 by sample number. With this linear relation we can now add
a model styling goal, such as smooth
by sample number. With this linear relation we can now add
a model styling goal, such as smooth
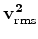 , given us the fitting goals
, given us the fitting goals
where 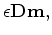 is the derivative operator,
is the derivative operator, 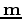 is the scaled
is the scaled
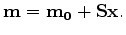 ,
and the model is
,
and the model is
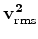 . We need a weighting term which gives higher
importance to good RMS picks, and equal weights all of the RMS velocities (undoing
the effect of the sample number scaling), resulting in
. We need a weighting term which gives higher
importance to good RMS picks, and equal weights all of the RMS velocities (undoing
the effect of the sample number scaling), resulting in
We can precondition the model by noting that
causal integration is the inverse of the derivative except at the first sample. We know
that first interval velocity value is the same as the first RMS velocity, resulting
in the final setting of fitting equations,
where  is the preconditioned variable and
is the preconditioned variable and 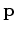 is a masking operator
that doesn't allow the first value to change. The advantage of selecting this
problem is that its solution is a rather thorough test of all the necessary features of
the solver. It involves a starting model, a weighting operator, and a masking operator.
It requires both chaining operators and making column operator objects. All of
the hard stuff is done away from the user in the solver, the user only needs
to construct the required operators and initialize and run the solver.
is a masking operator
that doesn't allow the first value to change. The advantage of selecting this
problem is that its solution is a rather thorough test of all the necessary features of
the solver. It involves a starting model, a weighting operator, and a masking operator.
It requires both chaining operators and making column operator objects. All of
the hard stuff is done away from the user in the solver, the user only needs
to construct the required operators and initialize and run the solver.
For this example the conversion is all handled in a module. The module begins by
using all of the modules that declare the operators and solvers, and the
declaration of variables.
module vrms_2_int_mod !Transform from RMS to interval velocity
use causint_mod !Causal integration
use weight_mod !Weighting operator
use mask_mod !Masking operator
use cg_step_mod !Conjugate gradient
use obj_solver_mod !Solver module
contains
subroutine vrms2int( niter, eps, weight, vrms, vint)
integer, intent( in) :: niter ! iterations
real, intent( in) :: eps ! scaling parameter
type(real_1d) :: vrms ! rms velocity
type(real_1d),target :: vint! interval velocity
real, dimension(:), pointer :: weight ! data weighting
integer :: st,it,nt
logical, dimension( size( vint%vals)) :: mask
logical, dimension(:), pointer :: msk
real, dimension( size( vrms%vals)) :: wt
real, dimension(:), pointer :: wght
type(prec_solver) :: p_s !Preconditioned solver
type(causint_op),target :: ca_op,ca2_op !Causal integration
type(mask_op),target :: m_op !Masking operator
type(weight_op),target :: wt_op !Weighting operator
type(cgstep),target :: cg !Conjugate gradient operator
type(real_1d),target :: dat !Data
Next we need to scale the data, create the weighting vector, and masking vector.
vrms2int.f90 vrms2int.unrat vrms_2_int_mod.mod
nt = size( vrms%vals)
call create_vec1(dat,vrms%vals)
do it= 1, nt
dat%vals( it) = vrms%vals( it) * vrms%vals( it) * it
wt( it) = weight( it)*(1./it) ! decrease weight with time
end do
mask = .false.; mask( 1) = .true. ! constrain first point
vint%vals = 0. ; vint%vals(1)= dat%vals( 1)
allocate(msk(size(mask)))
msk=.not.mask
allocate(wght(size(wt)))
wght=wt
Finally we need to initialize the operators, setup the solver,
and solve for the interval velocity squared.
call create_weight_op(wt_op,vrms,wght) !Create weighting op
call create_causint_op(ca_op,vint,"a1") !Causal op
call create_causint_op(ca2_op,vint,"a2")!Preconditioning
call create_mask_op(m_op,vint,msk) !Masking operator
p_s%lop=>ca_op; !Mapping operator
p_s%st=>cg; !Step function
p_s%pop=>ca2_op !Preconditioning operator
p_s%dat=>dat; !Data
p_s%mod=>vint !model
p_s%jop=>m_op; !Masking operator
p_s%wop=>wt_op !Weighting operator
p_s%eps=eps; !Scaling factor
p_s%p0=>vint !Initial preconditioned variable
call p_s%solve(niter) !Solve for interval v^2
call ca_op%op(.false.,.false.,vint,dat) !Estimated RMS^2
do it= 1, nt
vrms%vals( it) = sqrt( dat%vals( it)/it) !RMS velocity
end do
vint%vals = sqrt( vint%vals) !Interval velocity
deallocate(wght); deallocate(msk)
end subroutine
end module
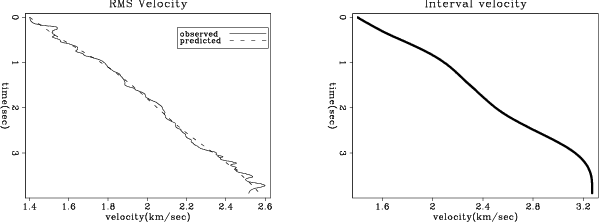
Figure 2 shows the result of running the inversion. The left panel
shows the original RMS velocity and the mapped RMS velocity. The right panel shows
the estimated interval velocity.
|
|---|
stiff
Figure 2. The left panel
shows the original RMS velocity and the mapped RMS velocity. The right panel shows
the estimated interval velocity.
|
|---|
![[pdf]](icons/pdf.png) ![[png]](icons/viewmag.png)
|
|---|
 |
 |
 |
 | Krylov space solver in Fortran 2009: Beta version |  |
![[pdf]](icons/pdf.png) |
Next: Conclusions
Up: Krylov space solver in
Previous: Solvers
2011-09-13
