The algorithm described in the previous sections will work well in the cases where there are few bad traces. In the cases where many traces are bad, this method will fail. One example of a failure would be where a single good trace falls between two bad traces.
For a shot gather with many bad traces, comparing more than just adjacent traces may provide more information than comparing only neighboring traces. For example, comparing each trace to the four nearest traces would allow a single trace between two bad traces to be marked as good, provided at least one of the four nearest traces is good enough to be used to predict the trace being considered.
Although a good trace might be better predicted from more than one or two neighboring traces, as more adjacent traces are used in the comparisons, the possibility of producing spurious good predictions increases. Even for the examples presented in the previous section, some samples in the bad traces remain, not because the samples are good, but because the bad trace happened to have a fairly low amplitude where a low amplitude was predicted. While these small errors are less harmful than the high-amplitude noise removed from the rest of the trace, removing them would be desirable. An obvious means of eliminating these spurious predictions would be to mute out a range of samples around any bad samples found. This approach is also appealing from a physical point of view, since any high-amplitude noise passing through the recording system is likely to have an lower-amplitude impulse response associated with it. Removing samples near high-amplitude noise could be considered as removing the impulse response of the noise.
Another approach to removing spurious predictions is to modify the comparisons of the residuals. For example, if four predictions are done, and three of the predictions indicate a sample is bad, the sample could be muted. In the previous work, any single good prediction is accepted. This idea of using the majority of the predictions to decide if a sample is eliminated will make the process more complicated and more time consuming than the previous methods, but may produce better editing in case where many of the traces are good. In the cases where many traces are bad, this method would require many more predictions than the technique demonstrated above.
Another alternative to the technique shown in the previous section would be extending the filters from
| |
(15) |
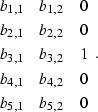 |
(16) |
![[*]](http://sepwww.stanford.edu/latex2html/cross_ref_motif.gif) .
A disadvantage of this approach would be that the traces must be fairly
clean before a good prediction is made.
The advantage of this approach is that it can be used to remove
noise with lower amplitudes and to remove traces with static shifts.
.
A disadvantage of this approach would be that the traces must be fairly
clean before a good prediction is made.
The advantage of this approach is that it can be used to remove
noise with lower amplitudes and to remove traces with static shifts.