




Next: Application to seismic
Up: Lomask and Biondi: Image
Previous: Introduction
The normalized segmentation method described by Shi and Malik (2000) is designed to look for clusters of pixels with similar intensity. To do this, it first creates a weight matrix relating each pixel to every other pixel within a local neighborhood. The strongest weights will be between pixels of similar intensity and close proximity to each other. The method then seeks to partition the image into two groups, A and B, by minimizing the normalized cut:
| 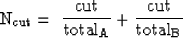 |
(1) |
where cut is the sum of the weights cut by the partition. totalA is the total number of weights in Group A, and totalB is the total number of weights in Group B. Normalizing the cut by the total number of weights in each group prevents the partition from selecting overly-small groups of nodes.
The minimum of Ncut can be found by solving the generalized eigensystem:
| 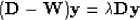 |
(2) |
created from the weight matrix (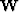 ) and a diagonal matrix (
) and a diagonal matrix (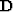 ), with each value on the diagonal being the sum of each column of
), with each value on the diagonal being the sum of each column of 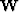 . The eigenvector (
. The eigenvector (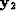 ) with the second smallest eigenvalue is used to partition the image by taking all values greater than zero to be in one group, and its complement to be in the other.
) with the second smallest eigenvalue is used to partition the image by taking all values greater than zero to be in one group, and its complement to be in the other.